AlphaFold vs. ESMFold: What Are the Differences?
AlphaFold and ESMFold: Two Ways to Predict Protein Structures
Predicting protein structure is an important step in biology. AlphaFold from DeepMind is famous, but Meta’s ESMFold is less known. What are the differences? Which one is better for your work? Let’s take a look.
1. Model Type: Language Model vs. Evolutionary Information
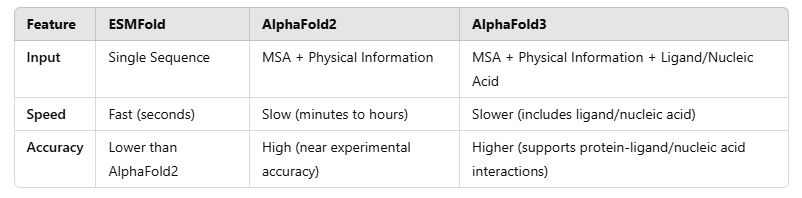
ESMFold: Made by Meta AI, ESMFold is based on ESM-2 (Evolutionary Scale Modeling 2), a big protein language model using Transformer. It predicts protein structure only from a single amino acid sequence, no need for evolutionary information (MSA).
AlphaFold: Made by DeepMind, AlphaFold uses deep neural networks (DNN) and graph neural networks (GNNs). AlphaFold2 needs multiple sequence alignments (MSA) and protein databases. It uses attention mechanism to find long-range residue connections. AlphaFold3 is even better, adding protein-ligand and protein-DNA/RNA interaction predictions.
When AlphaFold2 was released, we tried it on some proteins. Since it needs a huge MSA database, local installation was nearly impossible. We thought AlphaFold2 was just a bigger homology modeling tool. ESMFold, on the other hand, is a new idea, using language models to get protein embeddings.
2. Input Data: Single Sequence vs. Evolutionary Information
ESMFold is very fast because it only needs one sequence. It is useful for large-scale genome protein screening. AlphaFold2/3 is better for high-accuracy predictions, making them good for structural biology and drug design.
3. Speed: Which One Is Faster?
ESMFold is the fastest: No MSA calculation, can predict a structure in seconds.
AlphaFold2 is slower: Needs MSA search and deep learning, takes minutes to hours.
AlphaFold3 is even slower: Predicts protein-ligand and protein-nucleic acid interactions, needs more computation.
If you need fast predictions, ESMFold is better. If you need high accuracy, AlphaFold is the best. But in real work, extreme accuracy is not always needed, because molecular dynamics (MD) simulations can fix protein structures later.
4. Future Development
ESMFold: Meta closed the ESMFold team, but some people started a new company called EvolutionaryScale in New York. It is unclear if ESMFold will stay open-source.
AlphaFold: After winning the Nobel Prize, DeepMind will probably continue developing AlphaFold. But future open-source status is unknown.
5. Which One Should You Use?
Use Case: Fast single-sequence protein structure prediction (genome screening, etc.)
Best Model: ESMFoldUse Case: High-accuracy protein structure prediction
Best Model: AlphaFold2 (?) or ESMFold + MD simulationUse Case: Protein-ligand/nucleic acid interaction studies
Best Model: AlphaFold3
We believe ESMFold’s no-MSA method has a future. Protein information is much simpler than human language. If someone makes ESMFold smaller than 10B, it may be possible to fold a 100-residue protein in 10 seconds on a consumer GPU. Losing a little accuracy does not matter, because later processing will improve the structure.
Try ChemOrchestra™ (© 2025 QuantaBricks LLC, Patents Pending)
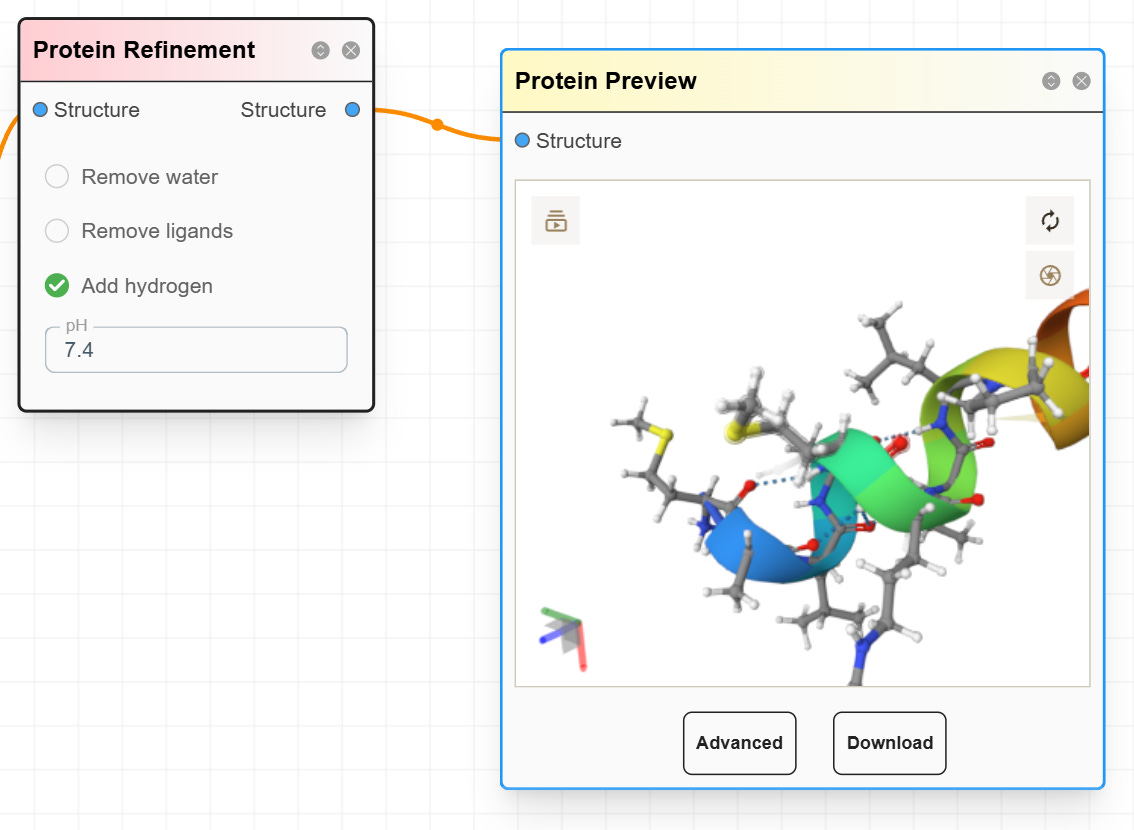
One problem with ESMFold is it does not add hydrogen atoms. Our ChemOrchestra™ platform can automatically add hydrogens and allow molecular docking. Future features will include structure optimization, solvation, and docking tools.
Try it for free! Interested? Leave a message or visit www.quantabricks.xyz.
For questions, contact our Product Manager at contact@quanta-bricks.com.