Get Started with Meta’s Molecular AI Model UMA on Google Colab
Meta and collaborators released the molecular foundation model UMA (Universal Model for Atoms) and its training dataset OMol25 (at ωB97M-V/def2-TZVPD level) this month.
You can easily experience the powerful UMA model directly in Google Colab, enabling fast and simple molecular adsorption simulations. This tutorial will walk you step-by-step from registering on Hugging Face to running UMA in Colab — beginner-friendly and straightforward!
Local installation is also extremely simple: just create a clean Python virtual environment and install with pip.
In recent years, big molecular models have been emerging rapidly. Notable examples include ANI-2x, AIMNet2, etc. However, for systems with fewer than 50 atoms (or ~1000 basis functions), these models offer little advantage — you’d better play with traditional DFT to avoid skeptical reviewers.
I believe the real value of molecular big models lies in the 50–3000 atom range, covering solid-state materials, metals, polymers, and large biomolecules. For even larger systems (>3000 atoms), traditional force fields remain the most practical solution due to computational speed.
Step 1: Register on Hugging Face and Request Access to UMA
Register a Hugging Face account
Visit the Hugging Face website and click “Sign Up” in the upper right. Fill in your email, username, and password.Request Access to UMA
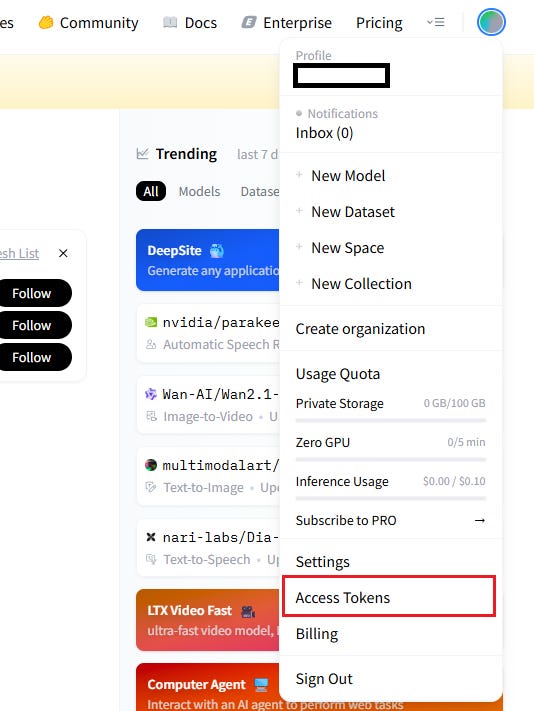
Go to the UMA model page, click to request access, and fill in a brief form. Approval is usually instant.Create an Access Token
Once logged in, click your avatar in the top right, select Access Tokens, and click Create new token. Copy and save your API key (e.g.,hf_xxxxxxxxxxxxxxxx) — you’ll need it soon.
Step 2: Set Up the Environment in Google Colab
Open Google Colab
Visit Google Colab and create a new notebook.Install Required Packages
Run the following in a Colab code cell:
!pip install ASE
!pip install git+https://github.com/facebookresearch/fairchem.git@fairchem_core-2.0.0#subdirectory=packages/fairchem-core Configure Your Hugging Face API Token
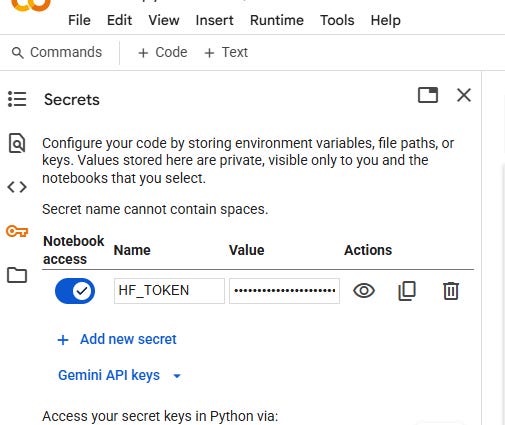
On the left Colab sidebar, click the 🔑 “Secrets” tab.
Click “Add a new secret”, name it
HF_TOKEN, and paste your Hugging Face token into the “Value” field.Once saved, Colab will load it automatically for model access.
Step 3: Run UMA with FAIRChemCalculator
UMA integrates seamlessly with ASE. Just use FAIRChemCalculator as your calculator. Here's an example of structure optimization:
from ase.build import fcc100, add_adsorbate, molecule
from ase.optimize import LBFGS
from fairchem.core import FAIRChemCalculator
from ase.io import read, Trajectory
# Initialize FAIRChemCalculator with UMA model
calc = FAIRChemCalculator(hf_hub_filename="uma_sm.pt", device="cpu", task_name="oc20")
# Load or build atomic structure
atoms = read('your_structure.vasp') # Replace with your VASP file path
atoms.calc = calc
# Set up trajectory output
traj = Trajectory('optimization.traj', 'w', atoms)
# Optimize using LBFGS
opt = LBFGS(atoms, trajectory=traj)
opt.run(fmax=0.05, steps=100)
# Convert trajectory to XYZ
frames = read('optimization.traj', index=':')
write('trajectory.xyz', frames)Note:
Replace
'your_structure.vasp'with your actual file.If you don’t have a structure file, you can build one using ASE’s tools:
atoms = fcc100('Pt', size=(2,2,3), vacuum=10.0) add_adsorbate(atoms, molecule('CO'), 1.5, 'ontop')
Step 4: Run Molecular Dynamics or Other Simulations
UMA works with ASE’s full simulation toolkit. Once your system is set up, you can run:
Geometry optimization (as shown above).
Molecular Dynamics using ASE’s built-in integrators:
from ase.md.velocitydistribution import MaxwellBoltzmannDistribution from ase.md.verlet import VelocityVerlet from ase import units MaxwellBoltzmannDistribution(atoms, temperature_K=300) dyn = VelocityVerlet(atoms, 5 * units.fs) dyn.run(1000)Code generation: You can ask ChatGPT to generate standard ASE code snippets based on your task and input structure.
Summary
By following the steps above, you can easily run UMA in Google Colab to simulate molecular adsorption, perform geometry optimization, or run MD simulations. I’ll be testing UMA on systems such as MOFs, heterogeneous catalysts, and solvated complexes in the future.
We’ve also provided the complete Jupyter Notebook, which you can import directly.
If you find this helpful, please star our GitHub repo so you won’t miss future updates.
Notebook link: https://github.com/myzzzz6/AI4Chem/blob/main/meta.ipynb
Important Notes:
Please do not use this model for publication before thorough benchmarking — avoid drawing potentially incorrect scientific conclusions.
Learning ASE will unlock many more simulation possibilities.
Got questions?
Post them at: https://www.reddit.com/r/ChemOrchestra/
(We'll reply in 24 hr)
Discord: https://discord.com/invite/A983ZhwwEY